Research at Barrow Neurological Institute
About Our Neuroscience Research
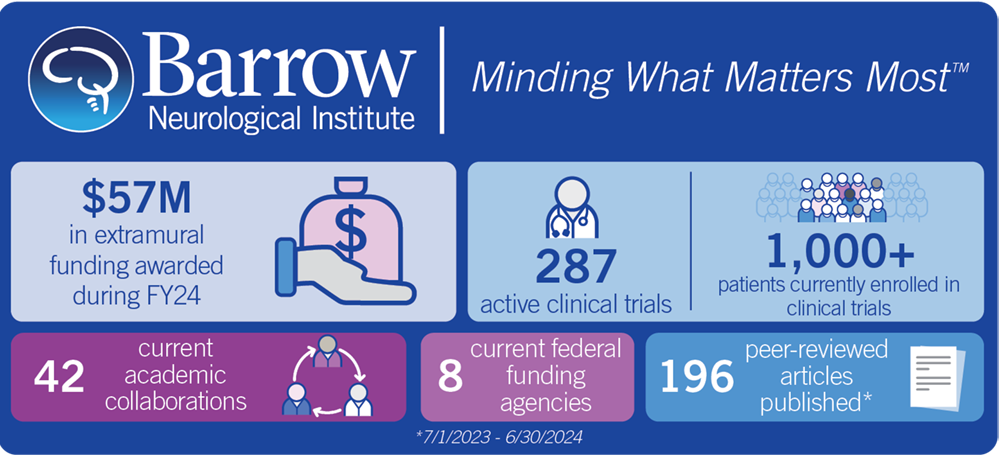
Neuroscientific research has been one of the three foundational pillars supporting the mission of Barrow Neurological Institute since our doors opened in 1962. What began as a handful of researchers and clinical trials decades ago has blossomed into a modern research powerhouse, with dozens of research scientists conducting translational work in the neurosciences, hundreds of clinical trials offering hope to patients with recalcitrant diseases, and numerous physician-scientists working to bring the latest discoveries from the laboratory to the clinic. We are also home to the Ivy Brain Tumor Center, the largest Phase 0 clinical trials program in the world.
Our physician-scientists in our Department of Neurology are conducting groundbreaking research in neuro-epidemiology, uncovering heretofore unknown risk factors for Parkinson’s disease, ALS, and Alzheimer’s disease.
Within the Department of Translational Neuroscience, Barrow scientists and physician-scientists conduct leading-edge research into neurodegenerative disease, including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, and Parkinson’s disease, as well as neuroimaging, traumatic brain injury, cerebrovascular diseases and stroke. They incorporate new and cutting edge technologies to explore disease pathogenesis as well as develop and test new therapies. We also have a strong history of innovation in neurosurgical devices and spinal instrumentation.
Our research program continues to specialize and adapt as advances are made in the field. We have a growing emphasis on the genetic underpinnings of diseases and pathologies like aneurysm, stroke, arteriovenous malformation, and brain tumors. In addition, our scientists are examining the epidemiological underpinnings of Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), and Parkinson’s disease.

Our Mission
To advance the knowledge and practice of medicine in neuroscience through basic and clinical research, education of medical professionals, and innovation in clinical techniques and technology.